SAQT-GWAS
Semi-Automated Quantitative Trait Genome-Wide Association Studies
DESCRIPTION
The SAQT-GWAS approach consists of a LONI Pipeline workflow and a collection of R functions that together allow a user to quickly and easily conduct GWAS on thousands of different quantitative traits simultaneously and subsequently visualize the results.
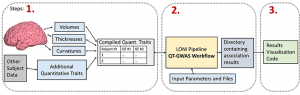
FEATURES
• Fast – 100s of QT-GWAS in ~10 minutes on the LONI cluster
• Flexible – use any set of quantitative traits as inputs
• Customizable – GWAS settings easily adjustable
• Simple – interrogate results with provided functions
• Powerful – enables systems-level exploration of genotype-phenotype associations
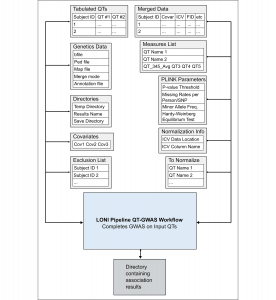
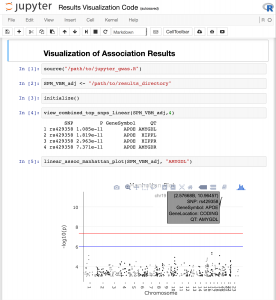
Download and Documentation
ACKNOWLEDGEMENT(S)
This work was in part supported by:
NIH grants P41EB015922 and U54EB020406.



